the Creative Commons Attribution 4.0 License.
the Creative Commons Attribution 4.0 License.
XLUM: an open data format for exchange and long-term preservation of luminescence data
Sebastian Kreutzer
Steve Grehl
Michael Höhne
Oliver Simmank
Kay Dornich
Grzegorz Adamiec
Christoph Burow
Helen M. Roberts
Geoff A. T. Duller
The concept of open data has become the modern science meme, and major funding bodies and publishers support open data. On a daily basis, however, the open data mandate frequently encounters technical obstacles, such as a lack of a suitable data format for data sharing and long-term data preservation. Such issues are often community-specific and best addressed through community-tailored solutions. In Quaternary sciences, luminescence dating is widely used for constraining the timing of event-based processes (e.g. sediment transport). Every luminescence dating study produces a vast body of primary data that usually remains inaccessible and incompatible with future studies or adjacent scientific disciplines. To facilitate data exchange and long-term data preservation (in short, open data) in luminescence dating studies, we propose a new XML-based structured data format called XLUM. The format applies a hierarchical data storage concept consisting of a root node (node 0), a sample (node 1), a sequence (node 2), a record (node 3), and a curve (node 4). The curve level holds information on the technical component (e.g. photomultiplier, thermocouple). A finite number of curves represent a record (e.g. an optically stimulated luminescence curve). Records are part of a sequence measured for a particular sample. This design concept allows the user to retain information on a technical component level from the measurement process. The additional storage of related metadata fosters future data mining projects on large datasets. The XML-based format is less memory-efficient than binary formats; however, its focus is data exchange, preservation, and hence XLUM long-term format stability by design. XLUM is inherently stable to future updates and backwards-compatible. We support XLUM through a new R package xlum, facilitating the conversion of different formats into the new XLUM format. XLUM is licensed under the MIT licence and hence available for free to be used in open- and closed-source commercial and non-commercial software and research projects.
- Article
(3691 KB) - Full-text XML
- BibTeX
- EndNote
Wilkinson et al. (2016) proposed four key principles for scientific data management towards open science: findability, accessibility, interoperability, and reusability – the FAIR guidelines. Since then, major funding bodies (e.g. Thorley and Callaghan, 2019; Agence Nationale de la Recherche, 2019; European Commission, 2021; Deutsche Forschungs Gemeinschaft, 2022) and publishers (e.g. Copernicus Publications, 2018; Wiley Author Service, 2022) have adopted these principles as part of their data management policies, and they have become an integral part of the European Code of Conduct for Research Integrity (ALLEA, 2017). If interweaved with umbrella terms such as “open data” or “open science”, the added value of transparency and reproducibility of modern science comes across as almost self-evident. Unfortunately, the implementation often seems to fall behind set goals. For instance, Perkel (2020) vividly covered the challenge of 35 participants trying to run decades-old computer code and concluded that maintaining reproducibility of software-based models and analysis pipelines over decades is a demanding, sometimes impossible, task. Likewise, we can infer that data formats tied to a small number of (outdated) programmes run the risk of data becoming inaccessible. Another aspect on the data side was considered by Noy and Noy (2020), who complained that common open data surrogate statements in articles such as data being “available upon request” may equate to no data access. Indeed, a pivotal aspect of the FAIR guidelines is their emphasis on principles fostering automated data processing or enabling such processing in the first place. The requirement to actively contact the study authors to request access to the data, e.g. e-mail requests, therefore inherently undermines the principles of open data (Noy and Noy, 2020). On the other hand, authors perhaps refrained from direct sharing because of unclear reporting guidelines or the effort required to document data with presumed low demand.
Adhering to the FAIR guidelines with actual benefits for all parties (e.g. data donators, data users, funding bodies) involves tackling low-level technical issues, such as defining an exchange data format enabling study authors to share their raw data in a manner which is structured, standardised, and ideally effortless, as well as in a format that will remain accessible long into the future. Here we adopt the idea that those issues are usually community-specific and best addressed through discipline-tailored solutions: for instance, for data generated in luminescence-based chronology studies.
Luminescence dating is a dosimetric dating method of key importance in Quaternary sciences and archaeology (e.g. Rhodes, 2011; Roberts et al., 2015; Bateman, 2019; Murray et al., 2021), covering around the last 300 000 years. In a nutshell, the datable event is the last sunlight or heat exposure of natural minerals such as quartz or feldspar. The dating process determines two parameters: (1) the absorbed dose (in Gy) accumulated in the minerals since the last heat or light exposure and (2) the environmental dose rate (in Gy ka−1). The ratio of dose (Gy) divided by dose rate (Gy ka−1) gives the age (ka). Methods frequently applied in luminescence dating studies are distinguished by their stimulation mode, e.g. thermally stimulated luminescence (TL; see Aitken, 1985), optically stimulated luminescence (OSL; Huntley et al., 1985), or infrared stimulated luminescence (IRSL; Hütt et al., 1988). Luminescence methods are also used by adjacent scientific disciplines, e.g. accident dosimetry and material characterisation (e.g. Yukihara and McKeever, 2011; Yukihara et al., 2014).
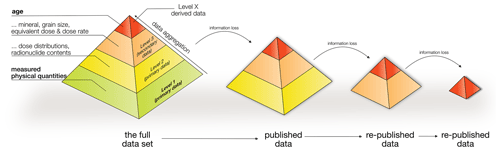
Figure 1A luminescence age is the result of data aggregation. In order to reproduce all steps, access to primary data (the base level) is indispensable. However, such primary data are seldom published or otherwise accessible. Republishing usually leads to information loss. The number of information and/or process levels in the graph is arbitrary.
Luminescence (dating) does not measure the absorbed dose directly but infers an equivalent dose (De) from the minerals' natural light output (luminescence) compared to a laboratory dose of known size. Luminescence dating studies and research building on such work routinely tabulate only a fraction of the recorded data in the form of aggregated parameters. One could think of a pyramidal information hierarchy with the age on the top (Fig. 1). The base is made out of minimally processed luminescence data, i.e. measured luminescence (for the purpose of this paper we “neglect” the dose rate information). Original dating studies ideally report the full information pyramid. However, the further an age is carried forward through subsequent studies or collected in data repositories, the higher the level of data aggregation. Good examples for aggregated luminescence data are repositories such as Lancaster et al. (2015) and Codilean et al. (2018). Such archives are excellent places to find locations of dating studies, but it is not easy to spatially link different ages without accessing the original studies with primary data.
Original, minimally processed luminescence data (see Fig. 1), i.e. measured luminescence, are hardly ever published along with a study. However, sharing of unprocessed luminescence data, accessible to others after the completion of a dating study, is desirable for several reasons.
-
Luminescence ages are end members of long measurement series involving various protocols, tests, and analysis steps with potentially different hardware and software tools. Once aggregated, it is challenging for others to revalidate published luminescence dates beyond plausibility checks. Shared raw data will potentially lead to better reproducibility and data quality.
-
Access to luminescence data on a single curve level supports the application of advanced analysis tools employing hierarchical Bayesian models such as the R package BayLum (Philippe et al., 2019) or the model “baSAR” (Combès et al., 2015; Mercier et al., 2016). Both approaches start with individual luminescence curves to integrate different parameters into a holistic model using Bayesian statistics to derive equivalent doses based on prior knowledge.
Other work has shown examples of how to study sediment pathways by tracing the bleaching histories of sediment grains (Chamberlain and Wallinga, 2019). If such data are never shared, their full potential remains untapped.
Recently, Balco (2020) advocated for a transparent and open middle-layer concept, disconnecting measured quantities from processed ages to account for changed, perhaps improved calculation procedures. His proposal was specific to cosmogenic nuclide exposure dating, but the general idea appears to be valid for other dating techniques, such as luminescence dating. For instance, it would enable others to test the impact of alternative applied statistical parameters on the calculated De in the future.
-
The approach of Balco (2020) renders ages moving targets; i.e. they may change with time due to different calculation procedures. Balco's approach emphasises the data treasure character of measured physical quantities (with “data are described with rich metadata”; FORCE11, 2014), which need to be preserved and shared instead of processed numbers. This approach holds for luminescence dating studies, which create, somewhat as a by-product, a vast amount of luminescence data on minerals from different origins. Such data are of potential interest, for instance, to geoscientists working on provenance analysis (e.g. Sawakuchi et al., 2018; Tsukamoto et al., 2011), to physicists focusing on luminescence models, or to data scientists trying to develop new approaches to enable exploratory luminescence data analysis to constrain physical parameters of OSL curves (e.g. Burow et al., 2016) or seeking training datasets to test machine learning approaches (e.g. Kröninger et al., 2019).
-
Broadly shared and accessible through a standard format, luminescence curve data will help establish a comprehensive repository for luminescence data, enabling studies and meta-studies not covered by the abovementioned examples.
Data sharing requests can only be reasonably accommodated if luminescence data can be easily exchanged, sufficiently archived, and analysed independently of proprietary software or file formats. We argue that one particular factor hampering the exchange and reuse of luminescence data is the absence of a suitable data format supporting long-term data preservation and fostering data exchange. To the best of our knowledge, long-term data preservation is an unresolved issue in the luminescence (dating) community. After being analysed and published, one can expect original primary data to be archived in compliance with scientific standards, but they may become inaccessible or incompatible with new data over time when the reanalysis is wanted. Such data are often lost to the public and need to be measured again. Hence, the first step of chronological data sharing and archiving is a data format that qualifies to serve that purpose.
In this contribution, we first briefly list existing data formats commonly used to store luminescence data. We then outline identified general technical requirements for a data format for the long-term preservation of luminescence data. Hereafter, we highlight features of a new XML-based file format, XLUM, developed for long-term preservation and exchange of luminescence data. The remainder provides examples and illustrates a reference implementation in R and Python, showing how existing data can be converted effortlessly into the new XLUM format. The discussion addresses potential shortcomings and challenges and canvasses future directions. We consider our contribution to be an initial definition, and the format blueprint is open to discussion within the luminescence community.
The XLUM file format is published and fully detailed at https://r-lum.github.io/xlum_specification (last access: 7 March 2023), a GitHub™ repository, and on Zenodo (European Organization For Nuclear Research and OpenAIRE, 2013; Kreutzer et al., 2022b). The chosen open-source MIT licence (https://opensource.org/licenses/MIT, last access: 2 May 2022) allows reuse in open-access and closed-source software projects.
In the remainder of this paper, we will use monospace letters for format and code snippets, as well as file format arguments. XML elements
(nodes), if not accompanied by a closing tag, are contracted into one short tag, for instance <node/> instead of <node> … </node>.
Equipment manufacturers have introduced most output data formats available in the luminescence dating community, for instance Daybreak (Bortolot, 2000), lexsyg (Freiberg Instruments, Richter et al., 2013, 2015), Risø TL/OSL reader (e.g. Bøtter-Jensen, 1988, 1997; DTU Nutech – Center for Nuclear Technologies, 2016), and SUERC-portable OSL (Sanderson and Murphy, 2010). Alternative formats were developed as part of research studies (e.g. Mittelstraß and Kreutzer, 2021). In other cases of equipment development, data output formats were not mentioned explicitly (Markey et al., 1997) or the hardware relied on export options of commercial laboratory software solutions (Guérin and Lefèvre, 2014; Mundupuzhakal et al., 2014). Some file formats are proprietary, and most are not documented in full. Additionally, data stored in comma-separated value files (file extension *.csv) or raster image-file formats (*.tif, *.spe) appear to be common; however, they lack the metadata required for luminescence data analysis.
Because (proprietary) file formats serve a particular purpose and equipment setup, the information stored in such files varies greatly across the different neatly tailored formats. Furthermore, they are incompatible in the sense that information transfer from one file format to another causes information loss (lossy coercion). The BIN/BINX format is very popular, which is well documented with good support from other manufacturers (e.g. Freiberg Instruments) and through community-maintained software solutions such as Analyst (Duller, 2015), LDAC (Liang and Forman, 2019), and the R (R Core Team, 2022) packages Luminescence (Kreutzer et al., 2012) and numOSL (Peng et al., 2013).
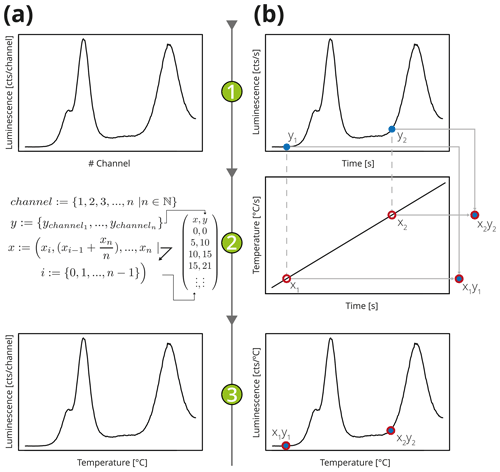
Figure 2Simplified illustration of two approaches to store a typical TL curve. (a) In the “conventional way” count data of the PMT are recorded channel-wise (1), and the temperature values are recalculated according to the equations in step (2) based on the minimum and maximum temperature values to obtain the (3) final TL curves, while data are imported in a programme. (b) In the approach suggested here, the luminescence signal and the temperature are recorded by two independent technical components, e.g. a PMT (1) and a temperature sensor (2) monitoring the heating process. While importing, the resulting TL curve (3) matches both recorded signals on the time domain. Such an import routine is e.g. available in the R package Luminescence (Kreutzer et al., 2012).
The BIN/BINX format was introduced decades ago, but it is not the most suitable candidate for long-term data preservation and exchange because of the following. (1) Different file format versions are incompatible because of non-identical file header lengths and byte order. (2) Storage of additional, so far unspecified, metadata requires a format change triggering a new format version. (3) For historical and memory-efficient reasons, instead of xy data, only y data (here counts per channel) are stored, and the temperature is deduced linearly from maximum and the minimum values (see Fig. 2). A thermoluminescence (TL) curve represents luminescence against stimulation temperature. However, a detection system usually consists of two independent technical components. One records the luminescence signal, e.g. a photomultiplier tube (PMT), and the other monitors the temperature, e.g. a thermocouple. Both quantities are recorded as a function of time, not temperature. (4) Data repositories should be findable (e.g. unique identifiers, proper metadata; see Wilkinson et al., 2016, Box 2, p. 4) and accessible by standard parser libraries (e.g. libxml), and the requirement of format-tailored software solutions should be avoided.
Mittelstraß and Kreutzer (2021)Table 1List of file formats dedicated to store luminescence data in alphabetic order (non-exhaustive).
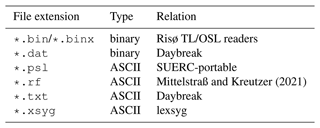
The other data formats listed in Table 1 suffer from similar or related problems because they were designed to accommodate data for a sole purpose or limited application range. In contrast, what is arguably preferable is a format that is as accessible and findable as possible and independent of a specific type of equipment – a requirement that laid the foundation for the development of XLUM.
A few design prerequisites guided the development of the XLUM format, and we list the most important below.
-
The format preserves physical quantities (measured, modelled), and their description remains equally readable to humans and machines.
-
Data are stored structured on a technical component and/or sensor level (e.g. photomultiplier, thermocouple) without limiting the data or forcing data reduction.
-
The format enables self-contained storage of data from technical components.
-
The format is self-explanatory; i.e. it can be generally understood without format documentation.
-
Backwards compatibility is maintained for future versions (newer format versions may carry additional attributes but remain readable to existing tools).
-
The format provides a neutral format that is open and “non-proprietary” with specifications defined by the scientific community not by equipment manufacturers.
-
The format application is permitted in closed and open software tools through suitable licence conditions.
-
Standard software solutions to process measurement data, e.g. MS Excel™, R, Python, LibreOffice, MATLAB™, and GNU Octave, are to be supported.
-
Data preservation and exchange are facilitated independently of the operating system running on users' personal computers.
-
The FAIR guidelines are supported by design and facilitate the creation of large repositories for long-term preservation and exchange of luminescence measurement and metadata.
We identified an XML (Extensible Markup Language)-based (W3C XML Core Working Group, 2008) format as the most suitable structure serving the outlined requirements.
The idea of introducing an XML-based format for storing luminescence data is not new. Bortolot and Bluszcz (2003) sketched a few general requirements for such a format 20 years ago, although this approach has not been widely adopted. An XML-based format is rather memory-inefficient, particularly if compared to binary formats, leading to relatively large files (tens of megabytes or more instead of megabytes). However, we believe that this aspect is of limited relevance because of the following. (1) Mass data storage is inexpensive, particularly if costs are compared to the year of Bortolot and Bluszcz (2003). (2) The overall amount of data produced in luminescence dating is negligible compared to other disciplines working with XML-based formats (e.g. Martens et al., 2011; Röst et al., 2015). (3) Modern storage systems of data repositories usually employ highly efficient low-level data compression methods (e.g. lossless data compression) independent of any file format, reducing the data footprint regardless of the exchange format.
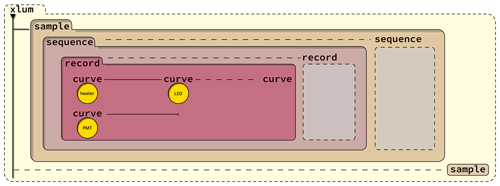
Figure 3The figure is a graphical representation of the data storage concept with the different node levels of the XLUM format. Data are stored sequentially over time. Dashed lines are used to indicate the possibility of multiple instances. For example, one XLUM file can contain many <xlum/> nodes, and one <xlum/> node may contain many <sample/> nodes.
In the following, we outline the conceptual structure of the XLUM format. To minimise verbosity, we only focus on key design concepts. For full details, we refer to our reference document on GitHub (https://xlum.r-luminescence.org, last access: 30 June 2022). The GitHub™ repository also contains a formal format description following the XML Schema Definition (XSD) for automated validation. XLUM defines a substructure, which can be part of a file or any other XML structure (W3C XML Core Working Group, 2008) that acts as a container or constitutes a file of its own, for instance with the file extension *.xlum, although the XLUM format does not enforce a specific file extension.
The two key features of the XLUM format are (1) information nesting, with measurement data only stored in the lowest node, and (2) support of data sharing by design.
4.1 Nesting of information on five node levels
The format consists of five levels (nodes) (Listing 1, Fig. 3), indicated by so-called tags. The correct formal description
requires an opening tag (<…>) and a closing tag (</…>) (see Listing 1). Each tag allows various
attributes (<tag attribute=” …>) for metadata, a few of which we will detail below. The number of attributes is not limited, and
additional user-defined attributes, not covered by the format definition, are explicitly allowed. However, the format definition insists on mandatory
attributes, with a few accepting the non-empty string NA for not available or not applicable. The first four upper nodes structure the data and
provide metadata to describe the dataset. The lowest node (<curve/>) contains the raw (or minimal processed) measurement data.
-
<xlum/>is the root node. It wraps all other data and is the parent to all other child nodes. The number of child nodes of<xlum/>is unlimited. Everything within one<xlum/>is considered a collection of data for different samples for which e.g. author names, a digital object identifier (DOI), and a licence can be assigned through attributes. -
<sample/>is the first child node to<xlum/>. It is the parent structure for luminescence data collected for a single sample. Hence, everything wrapped between<sample/>refers to a specific sample. Amongst others, expected attributes are thenameof the sample and the geographic coordinates (latitude,longitude). -
<sequence/>is the first child node to<sample/>. It sets the structure for measurement data defined through (measurement) sequences, e.g. a single aliquot regenerative (SAR) dose (Murray and Wintle, 2000) measurement sequence or any measurement data arranged in a particular order. Typical attributes areposition,fileName, orreaderName. -
<record/>holds all records of a sequence of a particular sample. A record is not necessarily limited to a single measured xy curve but a collection of xyz data created by technical components, which together define a record, e.g. a TL measurement. A typical argument isrecordType. -
<curve/>is the lowest node and the only node containing measurement data. All data stored in this node refer to a single technical component, e.g. a photomultiplier. One or many curves define one record. Numerical values in this node are separated by whitespace and span an array with three dimensions (see Eq. 1). Alternatively, this node allows data encoded as base-64 strings.
The crucial concept of the format is that data are stored only in <curve/> nodes, defined by technical components (actual or virtual)
measuring or simulating physical quantities over time. Data in <curve/> are numerical (measurement/simulation) values of a physical
quantity (discrete/continuous) spanning an array A of the form
with , where t is the extension of the array with respect to time (i.e. channels per
time instant), and x and y define the lateral geometry of the detector. Data are stored column-wise starting from before continuing in the time dimension. For instance, for a measurement over 100 channels with a photomultiplier
tube and . In contrast, for a measurement with a camera with a lateral resolution of
512 pixels × 512 pixels, , where remains
the same. The dimensional information is stored in the node attributes xValues, yValues, and tValues. All quantities,
except for t, are dimensionless (i.e. they have no default unit). However, the attributes xUnit, yUnit, and tUnit allow
setting SI units. For more attributes and their meaning, we refer to the detailed format description
(https://github.com/R-Lum/xlum_specification, last access: 4 July 2022).
To illustrate data storage in the XLUM format, we will pick one TL and one green stimulated luminescence (GSL) record belonging to a test
sequence measured for one sample. For simplicity, we limit the number of values for each curve to 10 and substitute NA with
“…” (three dots). We provide the complete file that can be imported correctly as a Supplement.
- Line 1
-
Mandatory entry, announcing the XML format. The format follows the Unicode® (The Unicode Consortium, 2022) UTF-8 and must not be changed. In a nutshell, it tells programmes for file parsing the character encoding and ensures that characters are interpreted correctly.
- Lines 2–5
-
Start of the XLUM record, with mandatory entries, e.g. for the namespace (
xmlns::…) and the format version used (here:1.0), as well as metadata related to the data, e.g.authorandlicense. Those attributes apply to all child nodes and clarify the data sharing rights in simple and unequivocal terms. In the example, we have applied the Creative Commons (CC) Licence CC-BY (https://creativecommons.org/about/cclicenses/, last access: 26 November 2022). This licence allows unrestricted data reuse, mixing, and sharing with the requirement to credit the data creators. - Lines 6–7
-
The
<sample/>node allows providing information about the sample, e.g.mineral,latitude. Those data are helpful for explorative data analysis with data from different geographical origins. - Lines 8–9
-
The
<sequence/>node, with general information that remains unchanged for a sequence, e.g.positionfor referring to a position in the equipment. - Line 10
-
The first record in the dataset, here of type
TL. - Lines 11–16
-
The complete entry for data as recorded by the thermocouple, i.e. the sensor monitoring the temperature of the period of 10 s (
tValues,tUnit) recorded in Kelvin (K,vUnit). The data (temperature values) are the content of the<curve/>node in line 15. For the other two curves, the measured values are listed in line 22 (PMT TL curve) and line 32 (count values GSL). - Lines 17–23
-
The second
<curve/>node with values measured with a PMT during heating (component). Here attributes fordetectionWindowandfilterare set. However, those attributes can be set toNAif information is not available or not applicable. - Lines 25–35
-
The second record in the sequence with one file with GSL measured with a PMT. While the record may contain information about the stimulation power or settings of the heating during stimulation, that information was not recorded for our example.
- Lines 35–37
-
The closing tags for the nodes
<sequence/>,<sample/>, and<xlum/>.
The example shows that a considerable part of the information refers to data giving information about the data measured with a technical component (PMT or thermocouple), rendering the dataset self-contained and portable without requiring additional documentation.
Beyond a community-tailored open data solution for storage and sharing, an open exchange format expands the availability of luminescence data beyond the target community. This enables experts from other disciplines to work with luminescence data. Data analytics is a field in the realm of artificial intelligence, machine learning, and data science. Here, two programming languages have gained significant popularity: R (R Core Team, 2022) and Python™ (Python Software Foundation, 2022). To foster the distribution and usage of XLUM, we developed two prototype interfaces: one for R and one for Python™.
6.1 Programming environment R
We realised support for the XLUM format for the statistical programming language R (R Core Team, 2022) through a new package called
xlum (Kreutzer and Burow, 2022). The package has reached a stable development status with a
test coverage of nearly 100 %; i.e. package functionalities are continuously tested in unit tests (see Kreutzer et al., 2017, for more
details) on different operating systems. Within the R environment, data elements can be accessed through base R functionality
and data types (e.g. list; see documentation of the packages xlum and
xml2).
In xlum, all file interactions (functions xlum::read_xlum(), xlum::write_xlum(),
xlum::validate_xlum()) are realised through the R package xml2 (Wickham et al., 2021)
interfacing the XML parser library libxml2 (https://gitlab.gnome.org/GNOME/libxml2, last access: 5 July 2022). The generation
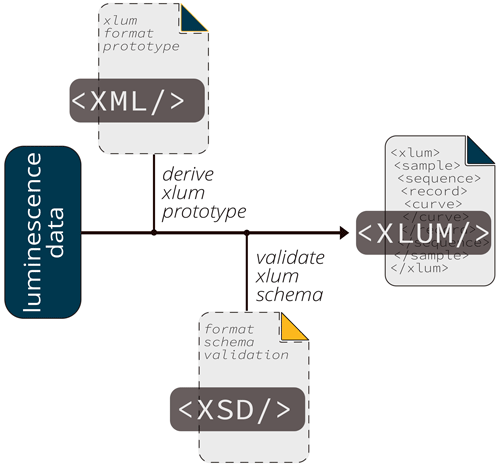
of XLUM files (function: xlum::write_xlum()) follows the workflow outlined in Fig. 4. First, a format blueprint is
derived from a prototype shipped with the package xlum. The prototype is then expanded and filled with
data. Before export, the file is validated (xlum::validate_xlum()) against an XSD schema to ensure that the produced XLUM
file follows the correct format specification. Both the prototype and the XSD are copies of files available as part of the XLUM
file format definition.
Other than basic import and export functionality to and from R of data shared as *.xlum files, the package
xlum supports the conversion of various file formats (e.g. *.binx, *.xsyg) to the
XLUM format proposed here using R functions commencing with convert_ (e.g. xlum::convert_binx2xlum() – see
Fig. 5). Internally, the conversion uses the R package Luminescence
(Kreutzer et al., 2012, 2022a) to create R-specific object structures (S4-class objects) tailored to luminescence data, called
RLum objects (see Kreutzer et al., 2017, their Fig. 2), and converts these structures using xlum::convert_rlum2xlum(). To date, this
conversion is not always lossless; i.e. not all metadata are transferred to the XLUM format for all formats due to the work-in-progress
character of the package xlum. We will improve the support with further maturity of XLUM. For
instance, the conversion of a *.binx file requires the R code lines in Listing 3.
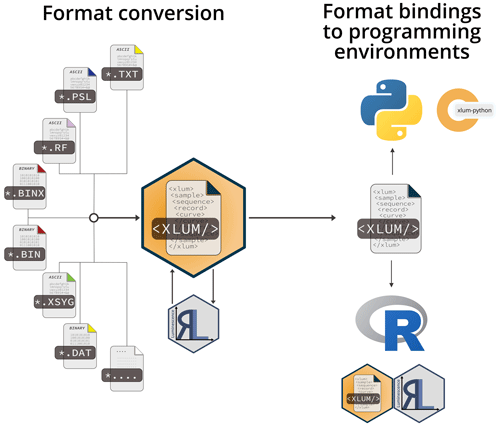
Figure 5The R package xlum supports the conversion of various commonly used luminescence (dating) data formats to <xlum/> using the R package Luminescence. Bindings to the statistical programming language R and the general-purpose programming language Python are realised through language-specific software packages.
6.2 Python
Similar to R, Python is an interpreted language. It is beginner-friendly and viral outside of traditional software development and computer science. A major advantage is the large and active open-source community maintaining a wide variety of packages (e.g. pandas, matplotlib, plotly) supporting data analysis workflows. For analysing luminescence data with Python, we provide a work-in-progress version of a package also called xlum via PyPI™ (https://pypi.org/project/xlum, last access: 8 January 2023). The package allows loading XLUM files with Python and conversion into pandas DataFrame objects (two-dimensional tabular data). This format is a starting point for further analysis, such as conversion to *.csv files, export to Microsoft Excel™, or graphical output. We show a minimalistic example of data import using Python in Listing 4. We provide more information and examples on the corresponding GitHub™ repository (https://github.com/SteveGrehl/xlum-python, last access: 1 July 2022).
Vor alles Figure 6 shows a simple representation of the measured values from an example file.
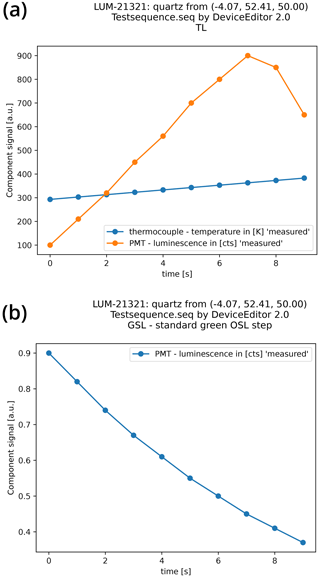
Figure 6Graphical representation of an XLUM example file, imported with the Python package xlum and visualised using the Python library matplotlib. Panel (a) shows a TL curve with data from two technical components (photomultiplier and temperature sensor), and panel (b) displays a so-called shine-down luminescence curve, i.e. photomultiplier data.
Our contribution aims to standardise luminescence data exchange and enable long-term data preservation. However, it is no attempt to bar or abolish other existing formats, which can often be considered primary data because conversion to XLUM may involve data coercion to some extent. However, the direct support of XLUM through other software and equipment manufacturers is desirable in the long run to make luminescence data more findable and accessible, promoting the FAIR data sharing guidelines. Nonetheless, it does not enforce them, and our contribution should not be understood as a claim on how and if data should be shared. Instead, here we refer readers to the guidelines of their institutes or funding bodies.
For XLUM, we have chosen an XML-derived format structure. It is usually less memory-efficient than any binary format, which we see as an acceptable weakness if it helps to facilitate human readability. During the specification process, we evaluated other similarly structured data exchange formats, such as JavaScript Object Notation (JSON) (https://www.json.org/, last access: 15 May 2023) and YAML (https://yaml.org, last access: 15 May 2023). In particular, discussions about the advantages and disadvantages of XML vs. JSON are considered on numerous IT websites, blogs, and platforms such as Stack Overflow (https://stackoverflow.com, last access: 15 May 2023) or in technology magazines. Without delving into their technical details, JSON has gained popularity over XML in recent years (e.g. Patrizio, 2016). Nevertheless, given the widespread use (e.g. Copernicus Publications, 2014) and support of XML schemas for data representation in various fields (see examples in Nolan and Lang, 2013), we opted for XML as a robust basis. XML provides standard grammar but remains flexible enough to be tailored to our purpose. If wanted and needed, luminescence data will remain easily transferable to other formats once standardised and archived as XLUM files. Another possibility is an amendment of XLUM, for instance, to facilitate better image data, for which storage is already possible today, and the optional support of base-64 string encoding enables a more efficient representation of those data.
Throughout the paper, we implicitly carried forward the limitation that the XLUM concerns luminescence data only, although a luminescence age is obtained from a luminescence-derived equivalent dose divided by a dose rate. Radionuclide concentration values used to calculate dose rates are derived using different methods, e.g. high-resolution γ-ray spectrometry, in situ γ-ray spectrometry, alpha or beta counting, and inductively coupled plasma mass spectrometry. Different methods make it challenging to develop a data format applicable to all methods. In contrast, nearly every luminescence dating laboratory has access to luminescence readers (recording luminescence data) with comparable technical capabilities. Those readers enable accurate and precise records of dim light emissions down to a single-grain level. Protocols and methods differ. The primary data are luminescence (light) in all cases. Still, a concept development, perhaps again focusing on luminescence dating, might be a reasonable attempt in the future.
The concept of open data has the notion of accessibility and data insight. However, shared data are not becoming automatically accessible, and not every dataset may provide similar valuable insight, but it depends on the experimental design. Making data accessible instantaneously with each study published appears to be advantageous for data users and disadvantageous for donors. For instance, Mills et al. (2015) discuss potential adverse effects on the availability of primary data in the field of biology if investigators of long-term studies are obliged to share their data. Although XLUM is merely a data format that sets no sharing rules, comparisons with different disciplines quickly wear thin. The fear of study authors that others may use their hard work to publish more quickly might be one of the reasons for the “upon reasonable request” data availability statements (Sect. 1). However, in the case of luminescence studies, long-term studies running over many years are scarce (e.g. Guérin and Visocekas, 2015, for an excellent example of such a study), and single datasets, even such as from a whole stratigraphic section as typical in palaeoenvironmental results studies, are of limited use to others. The true benefit of data sharing lies within many accessible and findable single datasets meaningfully linked through metadata, forming large datasets. With its component-focused design with minimum required metadata, XLUM does the groundwork for datasets aligned in luminescence-based chronologies across different sites in data mining projects concerned with luminescence model development and validation or in any explorative data analysis study.
Last, a significant obstacle to our initiative towards success is the question of broad community acceptance of the new format. Reasonable predictions are difficult to make. We tried to improve the chance of success of our initiative by implementing the first support in the programming language R and Python and by keeping all documents open-access. Furthermore, with the publication of this paper after peer review, the XLUM format will be supported by LexStudio2, the software running lexsyg luminescence readers, and could be supported by and/or adopted by other luminescence and dosimetry manufacturers. Further versions of this format will be developed transparently using the GitHub™ repository and are open to comments and contributions. Additionally, we propose allocating future format developments to a dedicated working group under the umbrella of a (to be formed) trapped-charge dating association.
Our contribution suggests an exchange and long-term data preservation format (XLUM) tailored to the specific requirements of the luminescence (dating) community. The format is XML-based and intended to store primary luminescence data and metadata self-consistently. The format implements (but does not enforce) the FAIR guidelines, facilitating a focus on accessibility and findability.
-
On the data storage level, XLUM does not constrain the amount of data stored for each measurement by an arbitrary format limitation; i.e. the number of components monitored is not limited by the file format. Furthermore, with this approach, the raw data are self-consistent and inherently contain all relevant information returned by a technical component.
-
On the data analysis level, the format design allows better data quality, as the data wanted for the analysis can be combined with additional information from other technical sensor data. These later data, e.g. stemming from a feedback system monitoring a particular instrument setting, might not be needed to answer the research question; however, it allows data validation and increases confidence in the result. For instance, failure of technical components may have invalidated the measurements and created artefacts. Such records can be excluded in the post-processing.
-
On the data exchange level, data can now be easily exchanged and combined, even though the file format version might be modified in the future, which might increase the overall transparency and value of measurement data.
-
On the preservation level, XLUM is suitable for long-term preservation of measurement data because it uses widely used and supported standard techniques (XML), and it is human-readable.
With XLUM, researchers can share their data using a unified file format or make them accessible via publisher websites to comply with various stakeholder guidelines, such as funding bodies. For the future, we hope that this approach can pave the way toward a new type of luminescence data repository, providing access to primary data on a single component level.
XLUM format specification: https://doi.org/10.5281/zenodo.7362438 (Kreutzer et al., 2022b). R package xlum: https://doi.org/10.5281/zenodo.7362364 (Kreutzer and Burow, 2022). Python package xlum: https://github.com/SteveGrehl/xlum-python (last access: 1 July 2022).
SK wrote the first draft of the paper, contributed to the original format specification, and developed and tested its implementation for R via xlum. SG led the original format design process and wrote down the first format specifications for the XSYG format. He later developed the Python application “xlum-python” to provide support for XLUM. MH contributed to the format specification and implemented its support as output format for the lexsyg TL and OSL readers. OS enhanced and refined the original format description. KD raised funds for the development of the predecessor XSYG 2013. GA added critical input to initial versions of this paper and its aims. CB validated and contributed to the xlum package. HR and GD contributed to the new format specification in the framework of the Marie Curie Action CREDit. All authors discussed and finalised the paper.
Freiberg Instruments GmbH is a manufacturer of commercial luminescence readers. Sebastian Kreutzer and Steve Grehl were employed at Freiberg Instruments GmbH when the XSYG file format, the predecessor of XLUM, was specified in early 2013. The initiative for this paper was undertaken independently by Sebastian Kreutzer in the framework of the Marie Curie Action CREDit to support the distribution of chronological reference data.
Publisher's note: Copernicus Publications remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
The XLUM file format has a predecessor, XSYG, originally designed in the course of a 2-month software project in early 2013. The two referees are thanked for their constructive comments on this paper.
This research has been supported by the Horizon Europe Marie Skłodowska–Curie Actions (grant no. 844457) (CREDit).
This paper was edited by Pieter Vermeesch and reviewed by Shannon Mahan and one anonymous referee.
Agence Nationale de la Recherche (ANR): Modèle de Plan de gestion des données (PGD), ANR, https://anr.fr/fileadmin/documents/2019/ANR-modele-PGD.pdf (last access: 24 April 2022), 2019. a
Aitken, M. J.: Thermoluminescence Dating, Academic Press, London, ISBN 978-0-12-046381-7, 1985. a
ALLEA: The European Code of Conduct for Research Integrity – REVISED EDITION, ALLEA, https://ec.europa.eu/info/funding-tenders/opportunities/docs/2021-2027/horizon/guidance/european-code-of-conduct-for-research-integrity_horizon_en.pdf (last access: 15 June 2022), 2017. a
Balco, G.: Technical note: A prototype transparent-middle-layer data management and analysis infrastructure for cosmogenic-nuclide exposure dating, Geochronology, 2, 169–175, https://doi.org/10.5194/gchron-2-169-2020, 2020. a, b
Bateman, M. D.: Handbook of luminescence dating, Whittles Publishing, Dunbeath, ISBN 978-184995-395-5, 2019. a
Bortolot, V. J.: A new modular high capacity OSL reader system, Radiat. Meas., 32, 751–757, https://doi.org/10.1016/s1350-4487(00)00038-x, 2000. a
Bortolot, V. J. and Bluszcz, A.: Strategies for flexibility in luminescence dating: procedure-oriented measurement and hardware modularity, Radiat. Meas., 37, 551–555, https://doi.org/10.1016/s1350-4487(03)00017-9, 2003. a, b
Bøtter-Jensen, L.: The automated Risø TL dating reader system, International Journal of Radiation Applications and Instrumentation, Part D, Nuclear Tracks and Radiation Measurements, 14, 177–180, https://doi.org/10.1016/1359-0189(88)90060-x, 1988. a
Bøtter-Jensen, L.: Luminescence techniques: Instrumentation and methods, Radiat. Meas., 27, 749–768, https://doi.org/10.1016/S1350-4487(97)00206-0, 1997. a
Burow, C., Zens, J., Kreutzer, S., Dietze, M., Fuchs, M. C., Fischer, M., Schmidt, C., and Brückner, H.: Exploratory data analysis using the R package 'Luminescence' – Towards data mining in OSL applications, https://doi.org/10.13140/RG.2.2.19673.62561, 2016. a
Chamberlain, E. L. and Wallinga, J.: Seeking enlightenment of fluvial sediment pathways by optically stimulated luminescence signal bleaching of river sediments and deltaic deposits, Earth Surf. Dynam., 7, 723–736, https://doi.org/10.5194/esurf-7-723-2019, 2019. a
Codilean, A. T., Munack, H., Cohen, T. J., Saktura, W. M., Gray, A., and Mudd, S. M.: OCTOPUS: an open cosmogenic isotope and luminescence database, Earth Syst. Sci. Data, 10, 2123–2139, https://doi.org/10.5194/essd-10-2123-2018, 2018. a
Combès, B., Philippe, A., Lanos, P., Mercier, N., Tribolo, C., Guérin, G., Guibert, P., and Lahaye, C.: A Bayesian central equivalent dose model for optically stimulated luminescence dating, Quat. Geochronol., 28, 62–70, https://doi.org/10.1016/j.quageo.2015.04.001, 2015. a
Copernicus Publications: Copernicus Press Release, Copernicus Publications provides full-text XML, Copernicus Publications, https://copernicus.org/news_and_press/2014-11-03_full-text-xml.html (last access: 30 May 2022), 2014. a
Copernicus Publications: Copernicus Press Release, 2018-11-05_Enabling-FAIR-Data-Commitment-Statement, Copernicus Publications, https://www.copernicus.org/news_and_press/2018-11-05_enabling-fair-data-commitment-statement.html (last access: 15 April 2022), 2018. a
Deutsche Forschungs Gemeinschaft (DFG): Guidelines for Safeguarding Good Research Practice: Code of conduct (v1.1), DFG, https://www.dfg.de/download/pdf/foerderung/rechtliche_rahmenbedingungen/gute_wissenschaftliche_praxis/kodex_gwp_en.pdf (last access: 24 April 2022), 2022. a
DTU Nutech – Center for Nuclear Technologies: The Sequence Editor, DTU Nutech, https://www.fysik.dtu.dk/english/research/radphys/research/radiation-instruments/tl_osl_reader/manuals (last access: 26 November 2022), 2016. a
Duller, G. A. T.: The Analyst software package for luminescence data: overview and recent improvements, Ancient TL, 33, 35–42, http://ancienttl.org/ATL_33-1_2015/ATL_33-1_Duller_p35-42.pdf (last access: 15 May 2023), 2015. a
European Commission: Data Guidelines | Open Research Europe, European Commission, https://open-research-europe.ec.europa.eu/for-authors/data-guidelines (last access: 10 June 2021), 2021. a
European Organization For Nuclear Research and OpenAIRE, Zenodo, https://doi.org/10.25495/7GXK-RD71, 2013. a
FORCE11: The FAIR Data Principles – FORCE11, https://force11.org/info/the-fair-data-principles/ (last access: 16 April 2022), 2014. a
Guérin, G. and Lefèvre, J.-C.: A low cost TL–OSL reader dedicated to high temperature studies, Measurement, 49, 26–33, https://doi.org/10.1016/j.measurement.2013.11.035, 2014. a
Guérin, G. and Visocekas, R.: Volcanic feldspars anomalous fading: Evidence for two different mechanisms, Radiat. Meas., 81, 218–223, https://doi.org/10.1016/j.radmeas.2015.08.009, 2015. a
Hütt, G., Jaek, I., and Tchonka, J.: Optical dating: K-Feldspars optical response stimulation spectra, Quaternary Sci. Rev., 7, 381–385, https://doi.org/10.1016/0277-3791(88)90033-9, 1988. a
Huntley, D. J., Godfrey-Smith, D. I., and Thewalt, M. L. W.: Optical dating of sediments, Nature, 313, 105–107, https://doi.org/10.1038/313105a0, 1985. a
Kreutzer, S. and Burow, C.: xlum: read, write, and convert XLUM data, Zenodo [code], https://doi.org/10.5281/zenodo.7362364, 2022. a, b
Kreutzer, S., Schmidt, C., Fuchs, M. C., Dietze, M., Fischer, M., and Fuchs, M.: Introducing an R package for luminescence dating analysis, Ancient TL, 30, 1–8, http://ancienttl.org/ATL_30-1_2012/ATL_30-1_Kreutzer_p1-8.pdf (last access: 15 May 2023), 2012. a, b, c
Kreutzer, S., Burow, C., Dietze, M., Fuchs, M. C., Fischer, M., and Schmidt, C.: Software in the context of luminescence dating: status, concepts and suggestions exemplified by the R package “Luminescence”, Ancient TL, 35, 1–11, http://ancienttl.org/ATL_35-2_2017/ATL_35-2_Kreutzer_p1-11.pdf (last access: 15 May 2023), 2017. a, b
Kreutzer, S., Burow, C., Dietze, M., Fuchs, M. C., Schmidt, C., Fischer, M., Friedrich, J., Mercier, N., Smedley, R. K., Christophe, C., Zink, A., Durcan, J., King, G. E., Philippe, A., Guérin, G., Riedesel, S., Autzen, M., Guibert, P., Mittelstrass, D., Gray, H. J., and Galharret, J.-M.: Luminescence: Comprehensive luminescence dating data analysis, CRAN, https://doi.org/10.5281/zenodo.6345291, 2022a. a
Kreutzer, S., Grehl, S., and Höhne, M.: XLUM data format specification: v1.0.0, Zenodo [code], https://doi.org/10.5281/zenodo.7362438, 2022b. a, b
Kröninger, K., Mentzel, F., Theinert, R., and Walbersloh, J.: A machine learning approach to glow curve analysis, Radiat. Meas., 125, 34–39, https://doi.org/10.1016/j.radmeas.2019.02.015, 2019. a
Lancaster, N., Wolfe, S., Thomas, D., Bristow, C., Bubenzer, O., Burrough, S., Duller, G., Halfen, A., Hesse, P., Roskin, J., Singhvi, A., Tsoar, H., Tripaldi, A., Yang, X., and Zárate, M.: The INQUA dunes atlas chronologic database, Quatern. Int., 410, 3–10, https://doi.org/10.1016/j.quaint.2015.10.044, 2015. a
Liang, P. and Forman, S. L.: LDAC: An Excel-based program for luminescence equivalent dose and burial age calculations, Ancient TL, 37, 21–40, 2019. a
Markey, B. G., Bøtter-Jensen, L., and Duller, G. A. T.: A new flexible system for measuring thermally and optically stimulated luminescence, Radiat. Meas., 27, 83–89, https://doi.org/10.1016/s1350-4487(96)00126-6, 1997. a
Martens, L., Chambers, M., Sturm, M., Kessner, D., Levander, F., Shofstahl, J., Tang, W. H., Römpp, A., Neumann, S., Pizarro, A. D., Montecchi-Palazzi, L., Tasman, N., Coleman, M., Reisinger, F., Souda, P., Hermjakob, H., Binz, P.-A., and Deutsch, E. W.: mzML—a Community Standard for Mass Spectrometry Data, Mol. Cell. Proteomics, 10, R110.000133, https://doi.org/10.1074/mcp.R110.000133, 2011. a
Mercier, N., Kreutzer, S., Christophe, C., Guérin, G., Guibert, P., Lahaye, C., Lanos, P., Philippe, A., and Tribolo, C.: Bayesian statistics in luminescence dating: The “baSAR”-model and its implementation in the R package “Luminescence”, Ancient TL, 34, 14–21, http://ancienttl.org/ATL_34-2_2016/ATL_34-2_Mercier_p14-21.pdf (last access: 15 May 2023), 2016. a
Mills, J. A., Teplitsky, C., Arroyo, B., Charmantier, A., Becker, P. H., Birkhead, T. R., Bize, P., Blumstein, D. T., Bonenfant, C., Boutin, S., Bushuev, A., Cam, E., Cockburn, A., Côté, S. D., Coulson, J. C., Daunt, F., Dingemanse, N. J., Doligez, B., Drummond, H., Espie, R. H. M., Festa-Bianchet, M., Frentiu, F., Fitzpatrick, J. W., Furness, R. W., Garant, D., Gauthier, G., Grant, P. R., Griesser, M., Gustafsson, L., Hansson, B., Harris, M. P., Jiguet, F., Kjellander, P., Korpimäki, E., Krebs, C. J., Lens, L., Linnell, J. D. C., Low, M., McAdam, A., Margalida, A., Merilä, J., Møller, A. P., Nakagawa, S., Nilsson, J.-Å., Nisbet, I. C. T., van Noordwijk, A. J., Oro, D., Pärt, T., Pelletier, F., Potti, J., Pujol, B., Réale, D., Rockwell, R. F., Ropert-Coudert, Y., Roulin, A., Sedinger, J. S., Swenson, J. E., Thébaud, C., Visser, M. E., Wanless, S., Westneat, D. F., Wilson, A. J., and Zedrosser, A.: Archiving primary data: Solutions for long-term studies, Trends Ecol. Evol., 30, 581–589, https://doi.org/10.1016/j.tree.2015.07.006, 2015. a
Mittelstraß, D. and Kreutzer, S.: Spatially resolved infrared radiofluorescence: single-grain K-feldspar dating using CCD imaging, Geochronology, 3, 299–319, https://doi.org/10.5194/gchron-3-299-2021, 2021. a, b
Mundupuzhakal, J., Adhyaru, P., Chauhan, N., Vaghela, H., Shah, M., Chakrabarty, B., and Acharya, Y.: FPGA based TL OSL system with EMCCD for luminescence studies, J. Instrum., 9, P04001, https://doi.org/10.1088/1748-0221/9/04/P04001, 2014. a
Murray, A., Arnold, L. J., Buylaert, J.-P., Guérin, G., Qin, J., Singhvi, A. K., Smedley, R., and Thomsen, K. J.: Optically stimulated luminescence dating using quartz, Nature Reviews Methods Primers, 1, 72, https://doi.org/10.1038/s43586-021-00068-5, 2021. a
Murray, A. S. and Wintle, A. G.: Luminescence dating of quartz using an improved single-aliquot regenerative-dose protocol, Radiat. Meas., 32, 57–73, https://doi.org/10.1016/s1350-4487(99)00253-x, 2000. a
Nolan, D. and Lang, D. T.: XML and web technologies for data sciences with R, Springer, https://doi.org/10.1007/978-1-4614-7900-0, 2013. a
Noy, N. and Noy, A.: Let go of your data, Nat. Mater., 19, 128–128, https://doi.org/10.1038/s41563-019-0539-5, 2020. a, b
Patrizio, A.: XML is toast, long live JSON, https://www.cio.com/article/238300/xml-is-toast-long-live-json.html (last access: 30 May 2022), 2016. a
Peng, J., Dong, Z., Han, F., Long, H., and Liu, X.: R package numOSL: numeric routines for optically stimulated luminescence dating, Ancient TL, 31, 41–48, http://ancienttl.org/ATL_31-2_2013/ATL_31-2_Peng_p41-48.pdf (last access: 15 May 2023), 2013. a
Perkel, J. M.: Challenge to scientists: does your ten-year-old code still run?, Nature, 584, 656–658, https://doi.org/10.1038/d41586-020-02462-7, 2020. a
Philippe, A., Guérin, G., and Kreutzer, S.: BayLum – an R package for Bayesian analysis of OSL ages: an introduction, Quat. Geochronol., 49, 16–24, https://doi.org/10.1016/j.quageo.2018.05.009, 2019. a
Python Software Foundation: Python Language Reference, Python Software Foundation, https://www.python.org (last access: 1 July 2022), 2022. a
R Core Team: R: A language and environment for statistical computing, https://r-project.org/ (last access: 30 April 2021), 2022. a, b, c
Rhodes, E. J.: Optically stimulated luminescence dating of sediments over the past 200,000 years, Annu. Rev. Earth Planet. Sc., 39, 461–488, https://doi.org/10.1146/annurev-earth-040610-133425, 2011. a
Richter, D., Richter, A., and Dornich, K.: lexsyg — a new system for luminescence research, Geochronometria, 40, 220–228, https://doi.org/10.2478/s13386-013-0110-0, 2013. a
Richter, D., Richter, A., and Dornich, K.: Lexsyg smart — a luminescence detection system for dosimetry, material research and dating application, Geochronometria, 42, 202–209, https://doi.org/10.1515/geochr-2015-0022, 2015. a
Röst, H. L., Schmitt, U., Aebersold, R., and Malmström, L.: Fast and Efficient XML Data Access for Next-Generation Mass Spectrometry, PLOS ONE, 10, e0125 108, https://doi.org/10.1371/journal.pone.0125108, 2015. a
Roberts, R. G., Jacobs, Z., Li, B., Jankowski, N. R., Cunningham, A. C., and Rosenfeld, A. B.: Optical dating in archaeology: thirty years in retrospect and grand challenges for the future, J. Archaeol. Sci., 56, 41–60, https://doi.org/10.1016/j.jas.2015.02.028, 2015. a
Sanderson, D. C. W. and Murphy, S.: Using simple portable OSL measurements and laboratory characterisation to help understand complex and heterogeneous sediment sequences for luminescence dating, Quat. Geochronol., 5, 299–305, https://doi.org/10.1016/j.quageo.2009.02.001, 2010. a
Sawakuchi, A. O., Jain, M., Mineli, T. D., Nogueira Jr, L., D. J. B., Häggi, C., Sawakuchi, H. O., Pupim, F. N., Grohmann, C. H., Chiessi, C. M., Zabel, M., Mulitza, S., Mazoca, C. E. M., and Cunha, D. F.: Luminescence of quartz and feldspar fingerprints provenance and correlates with the source area denudation in the Amazon River basin, Earth Planet Sc. Lett., 492, 152–162, https://doi.org/10.1016/j.epsl.2018.04.006, 2018. a
The Unicode Consortium: Unicode Technical Site, Unicode Consortium, https://www.unicode.org/main.html (last access: 23 May 2022), 2022. a
Thorley, M. and Callaghan, S.: NERC data policy – guidance notes (v2.2), Tech. rep., https://www.ukri.org/wp-content/uploads/2023/03/NERC-07032023-Guidance-notes-for-the-NERC-Data-Policy-final-22022023.pdf (last access: 15 May 2023), 2019. a
Tsukamoto, S., Nagashima, K., Murray, A. S., and Tada, R.: Variations in OSL components from quartz from Japan sea sediments and the possibility of reconstructing provenance, Quatern. Int., 234, 182–189, https://doi.org/10.1016/j.quaint.2010.09.003, 2011. a
W3C XML Core Working Group: Extensible Markup Language (XML) 1.0, 5th Edn., W3C, https://www.w3.org/TR/xml (last access: 14 June 2023), 2008. a, b
Wickham, H., Hester, J., and Ooms, J.: xml2: Parse XML, https://CRAN.R-project.org/package=xml2 (last access: 15 May 2023), 2021. a
Wiley Author Service: Open Data | Wiley, https://authorservices.wiley.com/open-research/open-data/index.html (last access: 16 April 2022), 2022. a
Wilkinson, M. D., Dumontier, M., Aalbersberg, I. J., Appleton, G., Axton, M., Baak, A., Blomberg, N., Boiten, J.-W., da Silva Santos, L. B., Bourne, P. E., Bouwman, J., Brookes, A. J., Clark, T., Crosas, M., Dillo, I., Dumon, O., Edmunds, S., Evelo, C. T., Finkers, R., Gonzalez-Beltran, A., Gray, A. J. G., Groth, P., Goble, C., Grethe, J. S., Heringa, J., t Hoen, P. A. C., Hooft, R., Kuhn, T., Kok, R., Kok, J., Lusher, S. J., Martone, M. E., Mons, A., Packer, A. L., Persson, B., Rocca-Serra, P., Roos, M., van Schaik, R., Sansone, S.-A., Schultes, E., Sengstag, T., Slater, T., Strawn, G., Swertz, M. A., Thompson, M., van der Lei, J., van Mulligen, E., Velterop, J., Waagmeester, A., Wittenburg, P., Wolstencroft, K., Zhao, J., and Mons, B.: The FAIR Guiding Principles for scientific data management and stewardship, Scientific Data, 3, 160018, https://doi.org/10.1038/sdata.2016.18, 2016. a, b
Yukihara, E. G. and McKeever, S. W. S.: Optically stimulated luminescence – fundamentals and application, John Wiley & Sons Ltd, West Sussex, United Kingdom, https://doi.org/10.1002/9780470977064, 2011. a
Yukihara, E. G., McKeever, S. W. S., and Akselrod, M. S.: State of art: Optically stimulated luminescence dosimetry – Frontiers of future research, Radiat. Meas., 71, 15–24, https://doi.org/10.1016/j.radmeas.2014.03.023, 2014. a
- Abstract
- Introduction
- Existing data formats in the luminescence dating community
- General data format requirements
- Format description
- Data representation: example
- Format support in R and Python
- Discussion
- Conclusions
- Code availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Financial support
- Review statement
- References
- Abstract
- Introduction
- Existing data formats in the luminescence dating community
- General data format requirements
- Format description
- Data representation: example
- Format support in R and Python
- Discussion
- Conclusions
- Code availability
- Author contributions
- Competing interests
- Disclaimer
- Acknowledgements
- Financial support
- Review statement
- References